Abstract
A pre-malignant phase known as IgM monoclonal gammopathy of undetermined significance (IgM-MGUS) precedes the development of Waldenström's macroglobulinemia (WM); an asymptomatic phase (aWM), which may also last for years, is followed by symptomatic WM which requires therapy. This transition from "pre-malignant” IgM-MGUS to overt WM is probably a multi-step process involving both clonal and microenvironmental changes. Current prognostic models for IgM monoclonal gammopathy risk stratification are mainly based on clinical variables and the underlying biology for the progression risk from the asymptomatic phases of the disease to symptomatic WM (sWM) remains unclear. The genetic composition of each clone is likely to determine the risk of disease progression, however, novel genetic prognostic markers are yet to be discovered. A detailed genetic landscape of "pre-malignant” lesions is missing and the aim of this study was to describe the genomic alterations that are present in IgM-MGUS and aWM patients and provide data to facilitate the discovery of potential genetic markers to improve current risk models.
A total of 60 patients with IgM monoclonal gammopathies were included in this study. This cohort included 33 patients with IgM-MGUS, 22 with aWM and 5 with symptomatic WM. All patients gave a written informed consent for sample collection and analysis. We performed whole-exome sequencing (WES) on 60 tumor samples of which 40 had matched tumor-normal samples and 20 were tumor-only samples, for a total of 100 samples. The analysis also included 3 serial samples from IgM-MGUS or aWM to sWM and 1 sample after treatment (within the 40 matched tumor-normal samples).
WES was performed on Illumina platforms with PE150 strategy, according to effective library concentration. Burrows-Wheeler Aligner (BWA) was utilized to map the paired-end clean reads to the human reference genome (hg38). For the SNP/InDel variant detection in the tumor samples the GATK (v4.0) pipeline was used. For the matched tumor - germline samples somatic SNP/InDel detection was performed with the MuTect/Strelka (v1.1.4 / v2.9.4) variant calling pipeline and for the somatic CNV detection the Control-FREEC (v11.4) software was used. The variants were annotated with ANNOVAR.
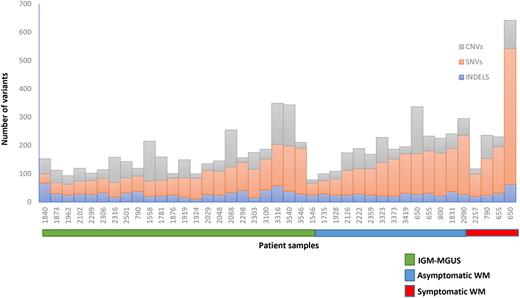
We observed an increasing tumor mutation burden (total number of mutations per patient) through the stages of disease evolution: the median number of single nucleotide variants (SNVs) in IgM-MGUS patients was 76 (range 32-161), in aWM was 115 (range 42-208) and in sWM was 213 (range 78-479) (Figure 1). Insertions-deletions (indels) and copy number variations (CNVs) remained similar among the 3 cohort of patients. From our previously described assay, MYD88L265Pwas detected in 48% of the of the IgM-MGUS patients, in 85% of the aWM patients and in 100% of sWM patients (Bagratuni et al. Blood Advances 2022). Among the 35 matched tumor-normal samples in IgM-MGUS and aWM patients, the most frequently mutated genes were IGLL5 (35%), NBPF1 (35%), MUC2 (30%) and KMT2C (23%). IGLL5 is among the recurrently mutated genes associated with progression to LPL/WM, a gene that plays a critical role in B-cell development. NBPF1 gene has been shown to be mostly mutated in follicular lymphoma patients specifically resistant to ibrutinib. MUC2 gene, among TP53 and MYC genes, has been reported in Burkitt lymphoma patients as being highly mutated through clonal evolution to relapsed disease while mutations in KMT2C gene impact epigenomic dysregulation which has been previously described in WM. Interestingly IgM-MGUS and aWM patients harboring MYD88L265P mutation had a significantly increased mutation burden (SNVs and CNVs) in other genes compared to patients with MYD88WTphenotype (p<0.05). Finally in patients with serial samples from aWM to sWM, one patient had a dramatic increase in the mutation burden from 143 to 479 SNVs which decreased after treatment to 120 SNVs, one patient had a slight increase from 150 to 163 SNVs while in one patient no alterations were observed. A more detailed catalog of the involved genes and analysis of additional samples will be presented at the meeting.
In conclusion, this is first study which focuses on the genetic profiling of early lesions (IgM-MGUS and aWM) in patients with IgM monoclonal gammopathies and suggests that increasing mutation burden could represent a potential biomarker for the identification of patients at high risk of progression.
Disclosures
Gavriatopoulou:Genesis Pharma: Honoraria; Sanofi: Honoraria; Karyopharm: Consultancy, Honoraria; GSK: Consultancy, Honoraria; Janssen Cilag: Honoraria; Takeda: Consultancy, Honoraria; Amgen: Consultancy, Honoraria. Terpos:Genesis: Honoraria, Research Funding; Amgen: Honoraria, Other: Travel expenses, Research Funding; BMS: Honoraria; EUSA Pharma: Honoraria, Other: Travel expenses; Sanofi: Honoraria, Research Funding; GSK: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Novartis: Honoraria; Takeda: Honoraria, Other: Travel expenses, Research Funding. Dimopoulos:BMS: Honoraria; Amgen: Honoraria; Beigene: Honoraria; Takeda: Honoraria; Janssen: Honoraria. Kastritis:Takeda: Honoraria; Pfizer: Consultancy, Honoraria, Research Funding; GSK: Honoraria; Genesis: Honoraria; Janssen: Consultancy, Honoraria, Research Funding; Amgen: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.